


|
Name |
Hao-Jen Huang |
|
Position |
Director, Department of Life Sciences; |
|
Telephone |
+886-6-275-7575 ext. 58126 |
|
|
|
|
Area of expertise |
Plant signal transduction, molecular biology, microbe engineering |
![]() Research Focuses: Stress Physiology
Research Focuses: Stress Physiology
![]() Research Aims
Research Aims
1. Stress signaling: systems biology to study plant responses to environmental stresses
The growth of plants is sessile, and plants cannot escape a stressful physical environment such as cold, heat, drought, heavy metal and interactions with insects or microorganisms such as fungi and bacteria. Like all living organisms, plants must sense the signals and respond to the external stimuli that influence their growth and development. In my lab, we used microarray and RNA seq assay to assess the effect of environmental stresses on the expression of plant genes. We also examined the possible involvement of ROS, calcium, CDPKs and MAPKs in stress signaling transduction pathways. Further characterization of the stress-responsive transcriptome and kinases signaling identified in my study will be of great value for improvement of plant breeding.
2. The role of microbes in improving crop health under stressful environments
How populations and species react to extreme environments is an important topic in ecology and evolution in the 21st century. Although there are several studies on the molecular base of how living organisms respond to extreme stress, the nature of plant adaptation to high stress habitats remains unresolved. Recently, several reports demonstrated that native plant species from high stress habitats require root-associated microbiomes for stress tolerance. In this research project, we will compare physiological, transcriptome and meta-transcriptome between stress tolerant populations and nearby normal control populations to gain insight into genomic, physiological, and biochemical mechanisms of evolved tolerance in two plants, which are currently unknown.
Using RNA-seq approach through the construction of root transcriptome and meta-transcriptome libraries, we will identify interlinked signaling pathways and biological process in response to extreme stresses. The outcome of the research project will lead to provide insights into the molecular mechanisms of plant responses and adaptations to extreme environment. Such information is essential to explore molecular mechanisms of environment stress response and evolved adaptive difference among wild population.
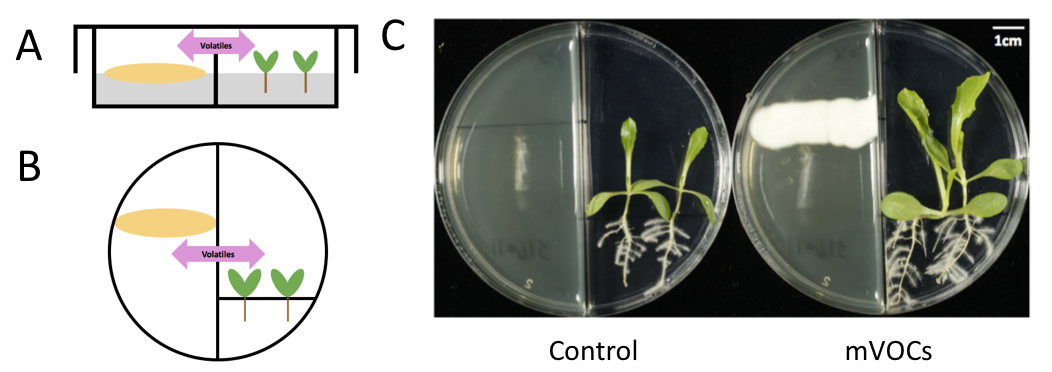
Fig. Microbial volatiles affect the growth of plants