Faculty: Ruey-Hua Lee Associate Professor

|
Name |
Ruey-Hua Lee |
|
Position |
Associate Professor |
|
Telephone |
+886-6-757575 ext. 58312 |
|
|
|
|
Area of expertise |
Plant Physiology, Cell biology, Molecular biology |
Education
|
University |
Country |
Major |
Degree |
Duration |
|
The University of London, Imperial College |
UK |
Biological Sciences |
PhD |
1993.10 – 1997.11 |
|
University of Bath |
UK |
Horticulture |
BsC |
1988.10 – 1991.05 |
Working Experiences
|
Research interests
![]()
I. Chloroplast degradation
We have identified a novel chloroplast α-galactosidase having enzyme activities only at alkaline pH. This enzyme (OsAkαGal) shows specificity to various α-1,6-galactosyl oligosaccharides and digalactosyl diacylglycerol (DGDG) containing non-reducing terminal α-galactosyl residues. Forward and reverse genetics show these plants have enhanced or delayed senescence speed, respectively (Figure 1). Over-expressed alkaline-α-galactosidase transgenic plant shows the assembly of thylakoid in the chloroplast completely distorted (Figure 2A-F). These plants also have poor growth than wild type and set seeds at 7 and 4 months, respectively. We conclude that DGDG is the substrate for OsAkαGal during thylakoid membrane disassembly. An Arabidopsis alkaline α-galactosidase (AtAKαGal3) has a similar function in regulating leaf senescence. Using thin-layer chromatography, we found this enzyme exhibits a dual function in RFOs degradation and synthesis of stachyose from raffinose. Structure analysis also shows that AtAKαGal has a catalytic and synthetic mechanism (Figure 3 A-C).
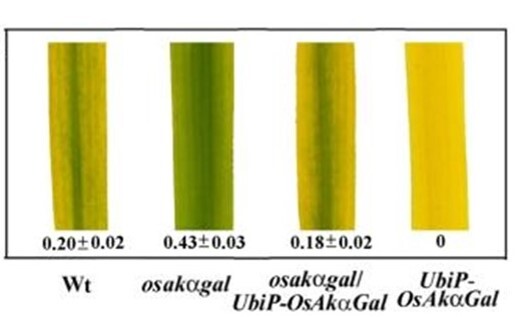
Figure 1. Ectopic expression of OsAkαGal affecting progression of leaf senescence.
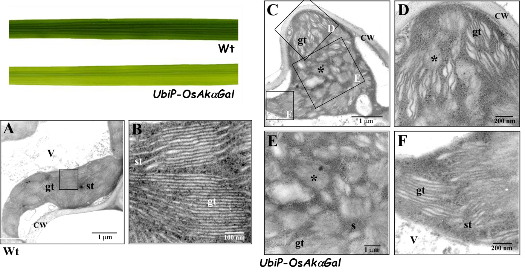
Figure 2. Over-expression of OsAkαGal affecting thylakoid membrane assembly in the chloroplast.
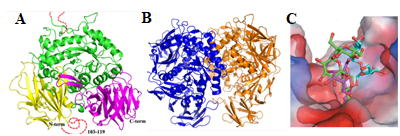
Figure 3. AtAkGal3 comprises three domains (A), Two AtAkαGal3 molecules (blue and orange) exist in one asymmetric unit (B),
Structure analysis shows AtAKαGal3 has catalytic and synthetic activities (C).
1. Lee R.H., Wang C.H., Huang L.T., Chen S.C.G. (2001) Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. Journal of Experimental Botany, 52:1117-1121.
2. Lee R.H. and Chen S.C.G. (2002) Programmed cell death during rice leaf senescence is nonapoptotic. New Phytologist, 155: 25-32.
3. Lee R.H., Lin M.C, Chen S.C.G.A. (2004) A novel alkaline α-galactosidase gene is involved in rice leaf senescence in rice. Plant Molecular Biology, 55: 281-295.
4. Lee R.H.*, Hsu J.H., Huang H.J. Lo S.F. and Chen S.C.G. (2009) Alkaline α-galactosidase degrades thylakoid membranes in the chloroplast during rice leaf senescence. New Phytologist 184: 596–606.
5. Chuankhayan P., Lee RH., ………Chen CJ. (2023) Structural insight into the hydrolase and synthase activities of an alkaline α-galactosidase from Arabidopsis from complexes with substrate/product. Acta Cryst. D79, 154–167.
![]()
II. Function of OsNACgs9 in vegetative and reproductive growth.
OsNACgs9 was identified using OsAkαGal promoter as bait and 1100 rice transcription factor library as prey in one-yeast-hybridization. We used Chimeric Repressor Gene-Silencing Technologyregenerated ZmUbiP::OsNACgs9-SDRX dominant suppression transgenic lines.. T2 transgenic plants have reduced tiller and panicle numbers (Figures A and B). In situ hybridization also found the transcripts mainly expressed in the ovule, stigma, and anther vascular tissues (Figure C). Percentages of empty seeds (red and blue in the bar chart) in these lines are 30-60% higher than in the wild-type (Figure D). Further studies are needed to validate the role of OsNACgs9 in regulating vegetative and reproductive development (Unpublish data).
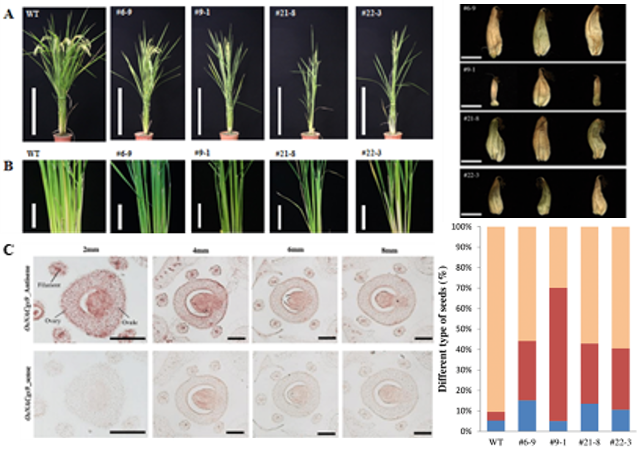
Figure 4. ZmUbiP::OsNACgs9-SDRX dominant suppression transgenic lines show reduced tiller (panicle) number (A, B),
OsNACgs9 strongly expressed in Ovary, anther filament and stigma tissues (C),
ZmUbiP::OsNACgs9-SDRX transgenic plants have reduced 30-60 % fully mature seeds compared to wild-type plants (D).
![]()
III. Investigate the functional role of an R2R3MYBS10-X transcription factor in thermotolerance during tomato reproductive development
Our studies show heat-sensitive CA4 show pre-mature cell death of tapetum and pollen mother cell during meiosis and tetrad stages of pollen development (Figure 5A). These defects were not observed in heat tolerant variety CLN1621L. Co-expression gene analysis discovered a link between genes involved in transcriptional cascade during pollen development and genes in phenylpropanoid flavonoid biosynthesis under heat stress (Figure 5B). We also uncovered a novel R2R3MYBS10-X transcription factor that may involve the regulation of phenlpropanoid/flavonoid biosynthesis. Ongoing molecular genetics and biochemical works are carried out to validate our hypothesis. CRISPR/Cas9 construct targets R2 and R3 MYB domains are used for genomic editing.
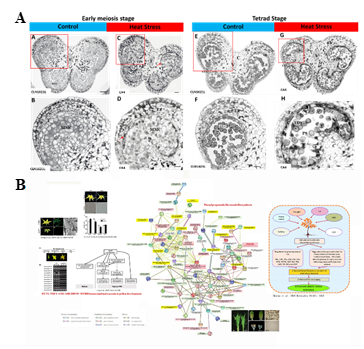
Figure 5. heat-sensitive CA4 shows pre-mature cell death of tapetum and pollen mother cell during meiosis and tetrad stages of pollen
development (A), Co-expression gene analysis discovered a link between genes involved in transcriptional cascade during pollen
development and genes in phenlpropanoid/flavonoid biosynthesis under heat stress (B).
![]() Journal and Conference Papers
Journal and Conference Papers
![]() Ministry of Science and Technology (MOST) Project
Ministry of Science and Technology (MOST) Project
