Faculty: Wen-Chi Chang Professor

|
Name |
Wen-Chi Chang |
|
|
Telephone |
+886-6-2757575 ext.58104 |
|
|
|
|
|
|
Area of expertise |
Bioinformatics, Genomics, Metagenomics |
|
|
Website |
|
![]() Position: Professor, Institute of Tropical Plant Sciences and Microbiology
Position: Professor, Institute of Tropical Plant Sciences and Microbiology
![]() Introduction of Laboratory
Introduction of Laboratory
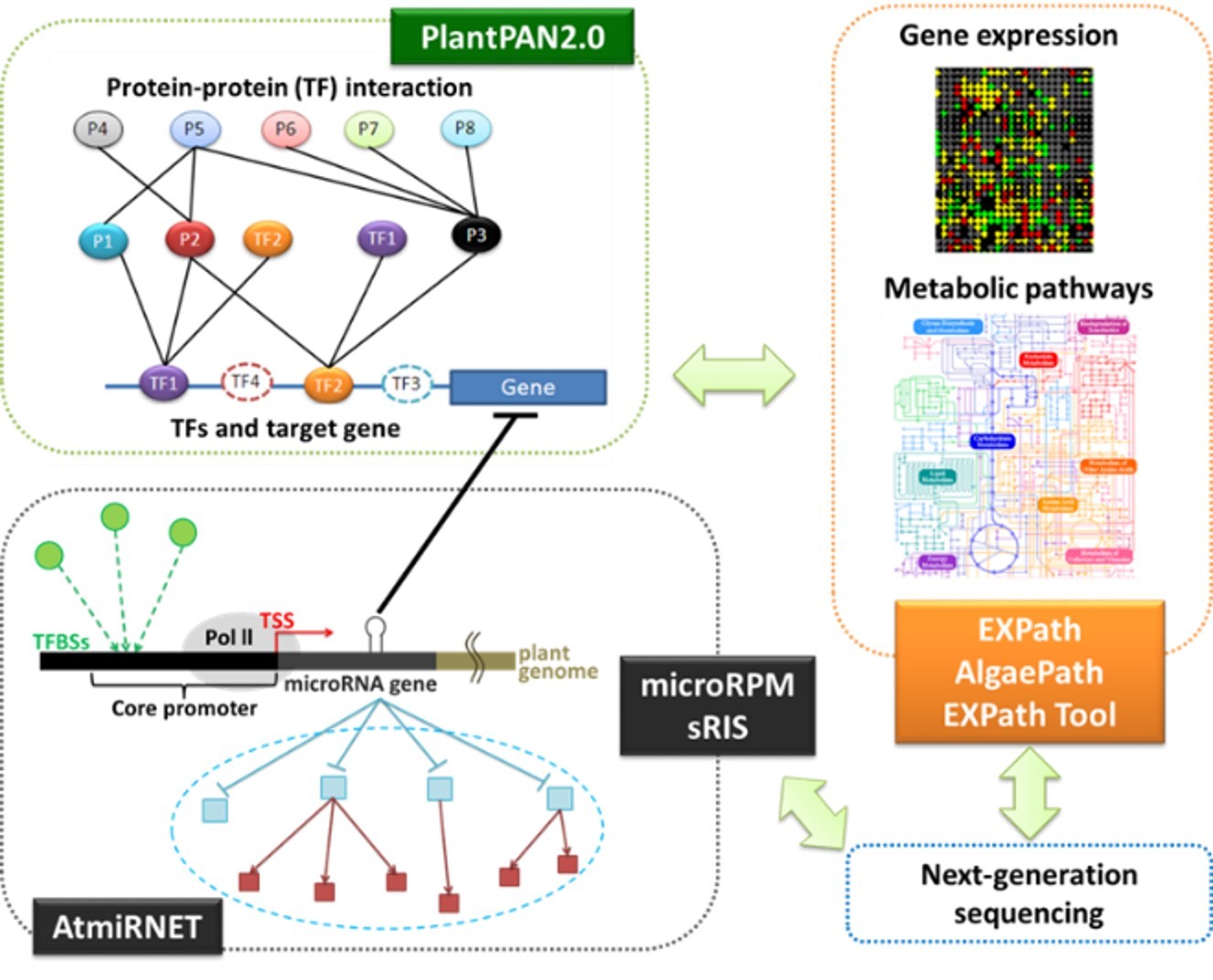
Our group actively develops plant bioinformatics systems, including PlantPAN2.0, EXPath, AlgaePath, EXPath Tool, AtmiRNET, microRPM, and sRIS for revealing gene regulatory mechanisms. The PlantPAN2.0 provides a systematic platform for the promoter analysis and reconstruction of transcriptional regulatory networks. The EXPath database was developed not only to comprehensively congregate the high-throughput gene expression data from over 1000 samples in biotic stress, abiotic stress, and hormone secretion, but also to allow the usage of this abundant resource for coexpression analysis and differentially expression genes (DEGs) identification. The AlgaePath is a web-based database that integrates gene information, biological pathways, and next-generation sequencing (NGS) datasets for the green algae. The EXPath Tool is an online software for the pathway annotation and comparative analysis of user-customized gene expression profiles derived from microarray or NGS platforms under various conditions to infer metabolic pathways for all organisms in the KEGG database. The AtmiRNET is a resource for reconstructing regulatory networks of Arabidopsis miRNAs. The microRPM is an effective prediction model to identify miRNAs from non-model plants based only on NGS datasets. Finally, the sRIS is a system to easily determine virus-derived small interfering RNA (vsiRNA), miRNA, and small RNA targets from NGS data of small RNA, degradome, and transcriptome. We hope the above resources can help scientists to clarify the gene regulatory mechanisms and discover critical marker genes for breeding from big bio-data. We also cooperate with Academia Sinica and Agricultural Research and Extension Station to study virus infection mechanisms in orchid and “plant probiotics” from soil, respectively.
![]() Research Aims
Research Aims
1. Construction of bio-system for plant transcription factors and gene regulatory networks
2. The interaction mechanisms between transcription factor and DNA sequence
3. Construction of the prediction model and regulatory networks for plant microRNAs
4. Comprehensive analysis of transcriptome and metabolome data
5. Identification of virus infection mechanisms in orchid
6. Metagenomics (data analysis and functional assay)
![]() Journal and Conference Papers
Journal and Conference Papers
![]() Ministry of Science and Technology (MOST) Project
Ministry of Science and Technology (MOST) Project
